Data cloud technology can accelerate FAIRification of the world’s biomedical patient data.
The growing field of precision medicine holds incredible promise for delivering better patient care and medical innovation, but there are barriers to greater implementation. As an emerging approach for disease treatment and prevention, precision medicine takes into account individual variability in genes, environment and lifestyle for each person. Its implementation has primarily been hastened by reducing sequencing costs. Next-generation sequencing (NGS) technology has dramatically dropped the price of genomic sequencing, from about $1 million in 2007 to $600 today per whole genome sequencing (WGS).
In medicine, lower sequencing costs and improved clinical access to NGS technology has been shown to increase diagnostic yield for a range of diseases, from relatively well-understood Mendelian disorders, including muscular dystrophy and epilepsy, to rare diseases such as Alagille syndrome. The data subsequently has expanded beyond NGS/sequences to multi-modal, which incorporates not only omics (proteomics, genomics, metabolomics) but also images and any other modalities that could be used in diagnosis, prevention and development of a cure for diseases in an effort to drive precision medicine at scale.
This approach allows doctors and researchers to predict more accurately which treatment and prevention strategies for a particular disease will work in which groups of people. It contrasts a one-size-fits-all approach, in which disease treatment and prevention strategies are developed for the average person, with less consideration for individual differences. In the context of life sciences, for example, analysis of tissue images for the presence of certain cell types in nuclei in cancer treatment aids as an important biomarker to understand disease progression and measure the efficacy of specific interventions.
The challenges: technical and ethical
With technological barriers for data capture and analysis no longer a concern thanks to the advent of the cloud, challenges remain primarily in two areas:
- Enabling ethical access with rigorous governance and privacy controls to the corpus of omics and allied data that has been acquired to date
- Performing tight integrations with clinical information of relevance, including study measurements, patient consent and patient characteristics
In most cases, the limiting factor for integration is the lack of a common identifier to link the data sets. In other instances, the concern is primarily the risk of potential patient re-identification that comes with longitudinal data enrichment. While the former can be solved by tokenization strategies provided by external vendors, the latter mandates the need for patient-level data enrichment to be performed with sufficient guardrails to protect patient privacy, with an emphasis on auditability and lineage tracking.
To summarize the importance of multiomics data analytics, the need of the hour is data governance around integrated data analysis that assures appropriate access, lineage tracking and auditability of analysis results.
In addition to the ethical challenges, there are other tactical issues while dealing with precision medicine data. Technology silos are often found within a single organization across different domains (research, clinical, medical) that generate these different modalities of the data (multiomics, images, clinical characteristics), and the complexity grows exponentially when considering that biomedical research is often conducted as collaborations across a host of private, public, academic and non-profit organizations.
For example, the data storage systems and processing pipelines that capture information from genomic sequencing instruments are very different from those that capture the clinical characteristics of a patient from a site. Also, the associated business metadata for omics, which make it findable for later use, are dynamic and complex and need to be captured separately. Additionally, the fact that they need to be standardized makes the data discovery effort challenging for downstream analysis. A sample representation of the business or functional metadata for omics type called RNA-seq is provided in Figure 1 below.
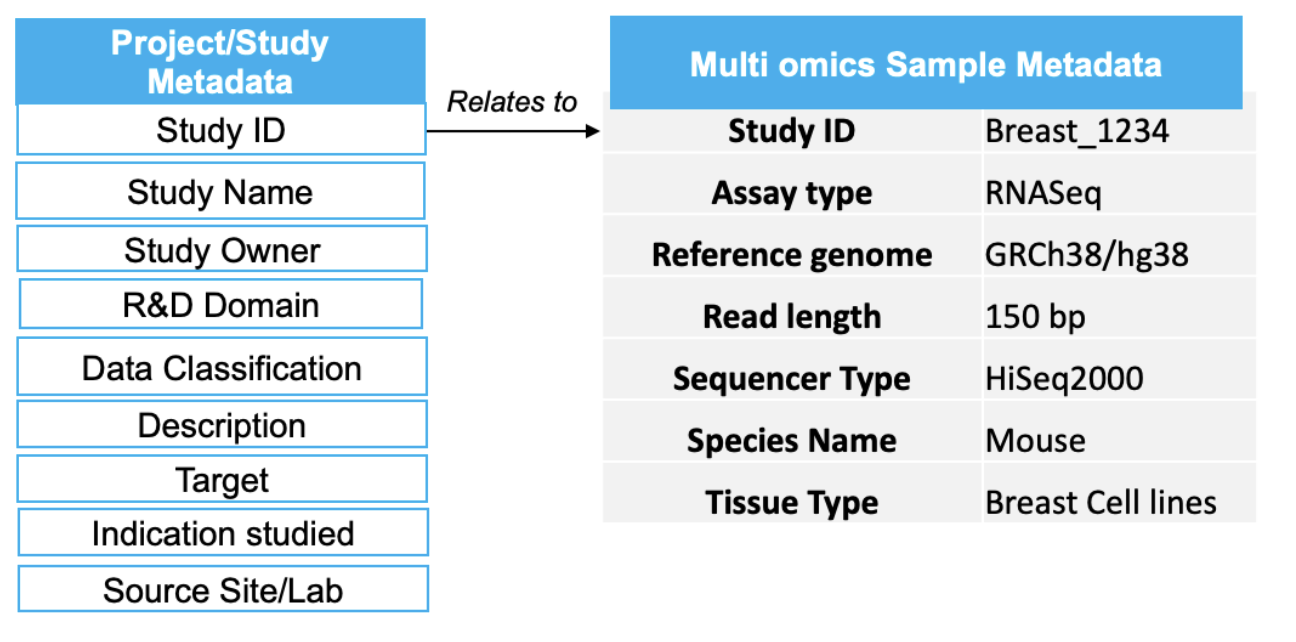
Figure 1: Sample metadata for omics that is needed for findability
This siloed data hampers effective exploratory analysis as a large amount of time is generally spent in identifying, collating and assessing the quality of data rather than in the analysis methodology. The data is there, it’s just not FAIR: Findable, Accessible, Interoperable and Reusable.
Defining FAIR data and it’s applications for life sciences
FAIR was a term coined in 2016 to help define good data management practices within the scientific realm. The principles emphasize machine-actionability (i.e., the capacity of computational systems to find, access, interoperate and reuse data with no or minimal human intervention) because humans increasingly rely on computational support to deal with data as a result of the increase in volume, complexity and creation speed of data. Delivering FAIR data requires the implementation of metadata and data models that strike a balance between usability and flexibility:
- Usability to ensure that consumers of the data can find and access the information that they require.
- Flexibility to ensure that the data itself is interoperable and reusable across varied research, clinical and real-world use cases.
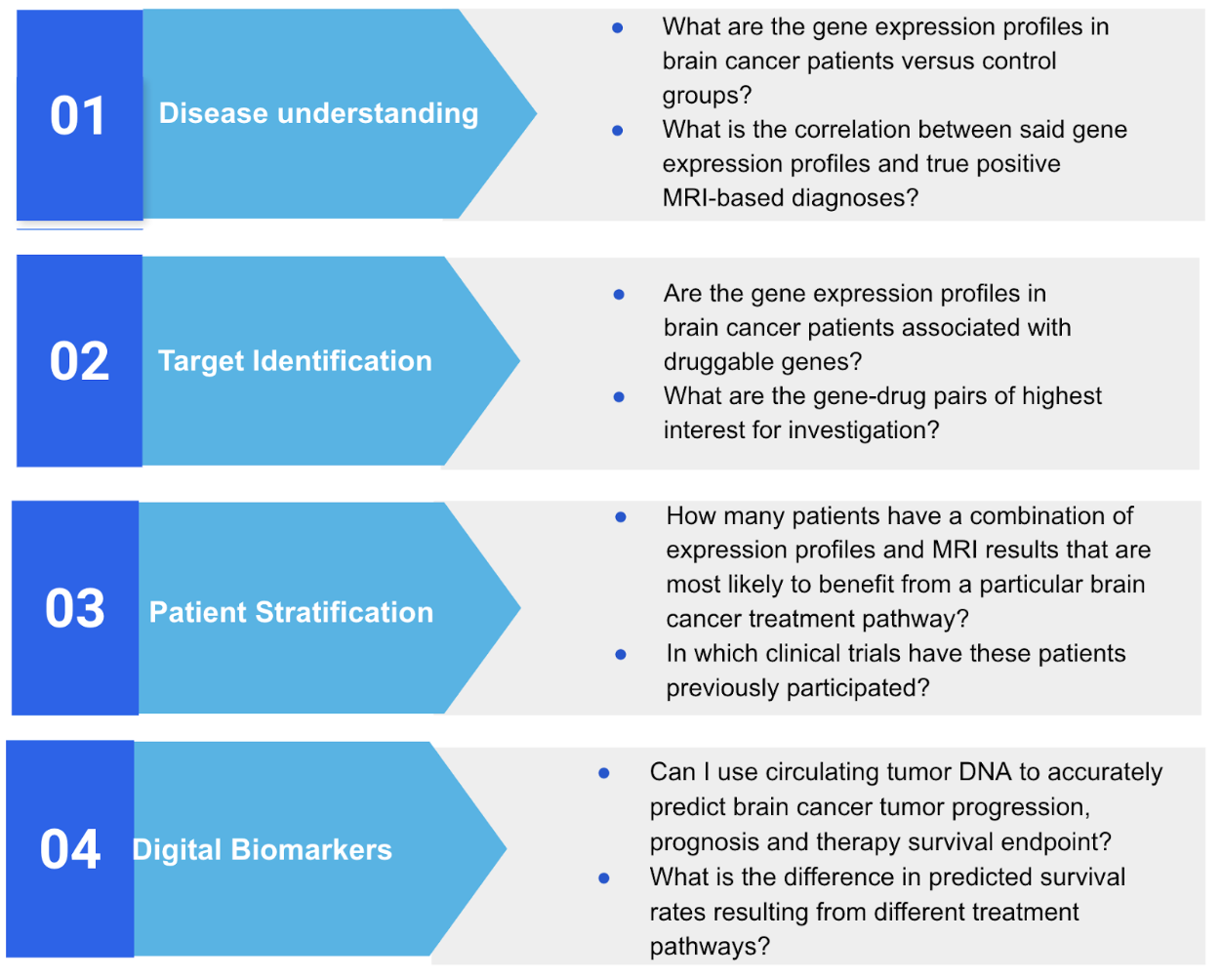
Figure 2: Questions answered by precision medicine
Snowflake and FAIR in the world of precision medicine and biomedical research
Cloud-based big data technologies are not new for large-scale data processing. However, new technological approaches have unleashed the ability to scale data processing and computing in a secure, seamless and reliable manner, and move compute closer to the data. A conceptual architecture illustrating this is shown in Figure 3.
Snowflake has a role to play in accelerating the creation of FAIRified data products, namely, data storage, data processing, collaboration and productization. They are well delineated in the table below:

Figure 3: Conceptual end-to-end system for enabling precision medicine with Snowflake.
Note: We have listed certain partner tools as an illustrative example. Snowflake connects to various third-party applications and allows seamless integration with a wide variety of tools.
Technological drivers
Data storage: Snowflake provides unprecedented flexibility to store a variety of data sources of all modalities (streaming, structured, semi-structured and unstructured) at a low cost, including omics data such as variant (VCF) data and unstructured data such as pathology images.
FAIR principle of Findable enabled by Snowflake:
- Allows users to build self-service analytics and insights via Streamlit apps and Snowsight to create findable applications, such as cohort explorers
- Natively integrates with third-party catalogs (e.g. Alation, Collibra) to some niche ones
- Allows easy ingestion of metadata (such as genomics metadata in Fig. 1) into the platform as a “variant column” that stores the free text, which can then be published into a Streamlit interface for searching. For example, in the case of DICOM images, metadata can be extracted and stored in a structured repository, which is then fed into a findability/cataloging tool.
Data processing: Snowflake provides a unified interface to explore and analyze data with SQL and Python/Java/Scala-based modalities, thus connecting both data scientists and data analyst personas in one ecosystem.
FAIR principle of Interoperable enabled by Snowflake:
- Supports multi-modal data ingestion like images, omics (VCFs), JSONs via native Snowpipe and Snowpark or via partner solutions
- Provides a schema discovery to infer schema without pre-imposing a data model natively
- Enables support for Iceberg Tables to aid interoperability across various standard open formats
Data collaboration: Snowflake’s collaboration mechanisms enable parties across organizations to securely share, access and analyze sensitive patient data in an ethical and compliant manner by providing role-based, row-level data access and masking policies. Biomedical research institutions can leverage data clean room technology to provide privacy-compliant Trusted Research Environments (TREs) at scale and at zero marginal cost to allow multiple collaborators to work together.
FAIR principle of Accessible enabled by Snowflake:
- Provides seamless and centralized governance to data with tight access controls natively from the platform
- Works in close collaboration with access provisioning tools such as Immuta
Data productization: Snowflake allows creation of intuitive digital experiences to create application-based data products such as Streamlit (Python-based framework).
FAIR principle of Reusable enabled by Snowflake:
- Accelerates collaboration with Snowflake Marketplace and private shares
- Provides access to the data at rest without data being moved
Enabling flexibility in data integration (across sources and formats), processing (across interface modalities), collaboration (across organizations) and productization without requiring the physical replication and movement of data, and connecting consumers and producers with minimal data movement, is a technology prerequisite for solving the FAIRification problem. At Snowflake, we envision a future in which private, public and non-profit healthcare and life sciences organizations leverage modern cloud data technologies to improve the accessibility of patient biomedical data, thereby accelerating our ability to understand and treat disease while conforming to the four principles of FAIR.
Snowflake is the pioneer of the Data Cloud, a global, federated network for secure, governed information exchange. Healthcare and life sciences organizations leverage Snowflake for their data management and support the FAIR needs in many different ways.
Learn more about how the Snowflake Healthcare & Life Sciences Data Cloud can help your organization manage your different modalities of data in precision medicine.
